· Jack Lee · Tutorials · 20 min read
Supercharging Medication NER with Local LLM and Two-Stage Retrieval RAG
Leveraging Local LLM to extract medical-related entities from unstructured prescription text with an advanced RAG architecture.

One of the common pain points of developing Named Entity Recognition (NER) models for domain-specific tasks has always been curating high quality training dataset, which would typically involve an exhaustive degree of human labor to manifest. Though with the advent of foundation models such as Large Language Models (LLM) that are capable of handling various generalized tasks, this alone has leveled the playing field quite considerably.
It is found that LLMs can be prompted to perform domain-specific tasks with pinpoint accuracy provided that proper context are supplemented, which is a boon for data curation involving both text annotation and entity extraction in the realm of NER. The benefits of such specialized LLMs are greatly magnified in the medical domain as healthcare professionals are stretched to perform various tasks in their day-to-day operations, leaving almost none to lend their assistance in the data curation process; these narrow-focused LLMs may serve as substitutes, with near human-level reasoning capabilities, to help provision high quality training dataset for Machine Learning (ML) models.
In this article, I will demonstrate how we could leverage LLMs to curate high quality training datasets for medication NER, utilizing a lightweight and efficient LLM such as Llama 3.2 3B, with a Two-Stage Retrieval RAG system that comprises of Hybrid Search and Reranking components to boost accuracy in extracting medical entities from unstructured text. While this system focuses on solving a specific problem, it lays the foundation for broader innovations in healthcare domain.
Check out the GitHub Repo for more details!
Table of Contents
The Problem Space
The unstructured medication or prescription text that this project hinges on, are derived from a set of synthetically generated electronic health records based on real-world statistical distribution; from an open source project called Synthea. Here are several examples of the unstructured medication text:
- Chlorpheniramine Maleate 2 MG/ML Oral Solution
- 1 ML Epoetin Alfa 4000 UNT/ML Injection [Epogen]
- Acetaminophen 325 MG / Oxycodone Hydrochloride 10 MG Oral Tablet [Percocet]
- 120 ACTUAT fluticasone propionate 0.11 MG/ACTUAT Metered Dose Inhaler [Flovent]
- 168 HR Ethinyl Estradiol 0.00146 MG/HR / norelgestromin 0.00625 MG/HR Transdermal System
It may seem complicated at first glance but there is a common pattern that can be gleaned from; that being drug name, dosage, quantity, administration type, and brand.
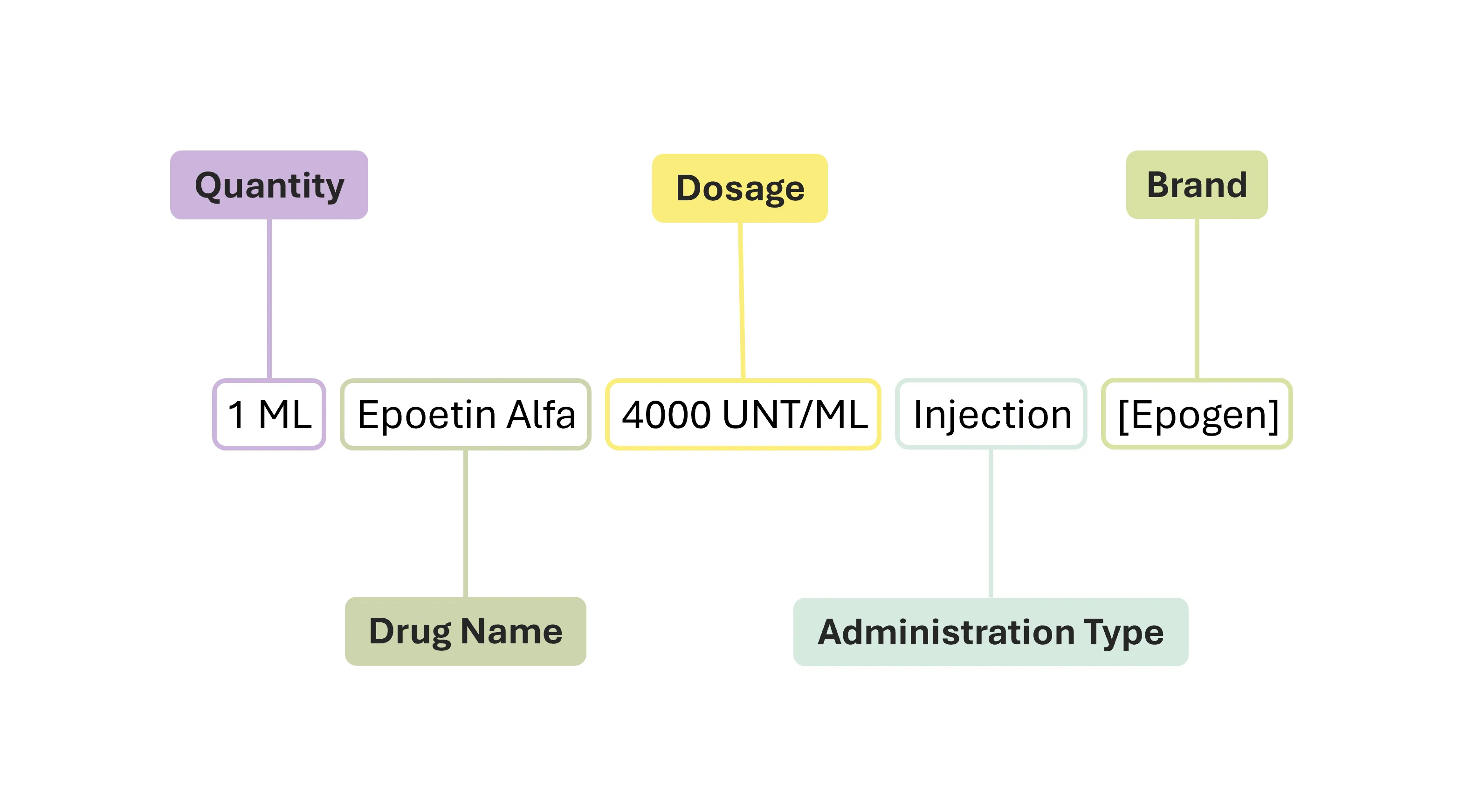
Consider the 2nd element of the list as the exemplar to be processed and entity extracted, it would likely assume the following JSON format:
{
"original_text": "1 ML Epoetin Alfa 4000 UNT/ML Injection [Epogen]",
"quantity": ["1 ML"],
"drug_name": ["Epoetin Alfa"],
"dosage": ["4000 UNT/ML"],
"administration_type": ["Injection"],
"brand": ["Epogen"]
}
Solution Overview
A Retrieval-Augmented Generation (RAG) system that combines Hybrid Search and Reranking will be able to handle unstructured data effectively. Here’s how this solution works:
- Hybrid Search: Retrieves relevant data chunks from a knowledge base using both dense and sparse embedding models for wider coverage of possible candidates.
- Reranking: Orders retrieved chunks based on their relevance.
- NER with LLMs: Uses Llama 3.2 3B to extract entities from the input query of medication text with the retrieved-reranked chunks as context.
Dense embeddings serve as a semantic filter to ensure that retrieved candidates are meaningfully related to the query, considering context and synonymy, while sparse embeddings serve as a lexical filter to ensure precise matches on specific terms, numbers, or keywords. This, in turn, allows the hybrid retriever to fetch candidates with higher precision.
The reranking component applies a computationally intensive scoring model such as cross-encoders to rank the retrieved candidates based on their relevance to the query, typically in a pairwise fashion with similarity scores.
The models chosen for the hybrid search and reranking are based on the criteria of being small and efficient, thus processing time are drastically reduced:
- Dense Embedding – “BAAI/bge-small-en-v1.5”
- Sparse Embedding – “Qdrant/bm42-all-minilm-l6-v2-attentions”
- Reranking – “cross-encoder/ms-marco-MiniLM-L-6-v2”
Llama3.2, with just 3 billion parameters, was chosen for its balance of efficiency and accuracy. Hosting it locally with Ollama ensures data privacy and eliminates reliance on cloud infrastructure, making the solution cost-effective and secure.
Building The Application
Experimental Phase
Prompt engineering was used as the initial litmus test to examine if local LLMs such as Mistral 7B and Llama 3.1 8B are truly capable of handling NER tasks in entity extractions with several few-shot examples to output valid JSON format. Lo and behold, it works, though sparingly; the accuracy tends to drop off when complex prescription texts are encountered, which is resolved by providing more diverse few-shot examples in the same context window, but this resulted in higher latency from the LLM response time as there are more tokens to be processed. The limitations of utilizing prompt engineering alone for handling NER tasks are quite apparent, each of which requires specific nuances to tackle:
- Few-shot Examples
- Must be relevant and concise to the input query.
- A dynamic mechanism should be in place to fetch the relevant examples.
- The fetched examples should be filtered for relevancy.
- Response Time Latency
- Smaller overall input context length allows faster response time.
- A smaller but efficient LLM should be used to reduce latency further.
Based on the lessons learned, a RAG system seems to be the most appropriate choice for tackling the issues with few-shot examples and response time latency, where the examples are stored in a vector database and a leaner language model being selected for inference.
Development Phase
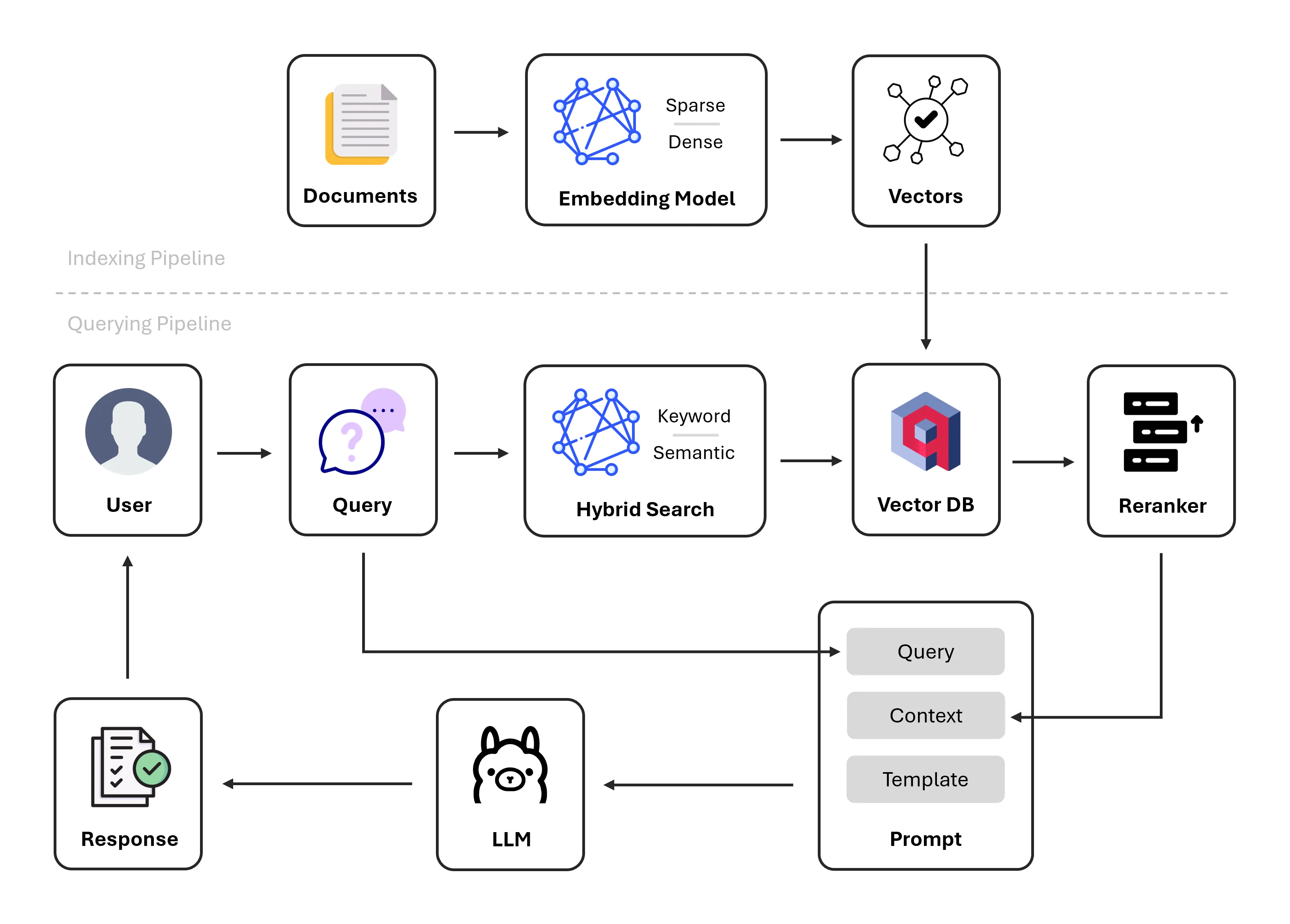
RAG Framework
After much experimentation with leading frameworks (E.g. LangChain, LlamaIndex, Haystack) for building RAG applications, Haystack was ultimately chosen; as it has one of the best documentations and straightforward API interfaces, to orchestrate both the indexing and querying pipelines. Custom logic can be easily integrated into Haystack components and plugged into existing pipelines, plus there are multiple cookbook tutorials to draw inspiration from.
It is important to note that the components of a Haystack Pipeline such as document store, embedders, reranker, prompt builder, etc cannot be shared between pipelines and thus new pipelines much be created for processing new inputs. This constraint alone has led to the adoption of the factory design pattern for instantiating pipelines to handle both indexing and querying mechanisms, especially useful when it functions within a FastAPI wrapper to handle POST requests concurrently. Here’s a code snippet of the pipeline factory, with all the class methods hidden away for brevity:
# app/core/pipeline/factory.py
import asyncio
from functools import partial
from haystack import Pipeline
from app.config.logging import get_logger
logger = get_logger(__name__)
class PipelineFactory:
async def create_indexing_pipeline(self) -> Pipeline:
"""Create indexing pipeline with concurrent component initialization"""
logger.info("Creating indexing pipeline...")
try:
# Initialize document store and both document embedders concurrently
doc_store, (dense_embedder, sparse_embedder) = await asyncio.gather(
self._async_init(self._create_doc_store),
self._async_init(self._create_document_embedders),
)
# Initialize document writer after we have the doc_store
document_writer = await self._async_init(
partial(self._create_document_writer, doc_store)
)
indexing = Pipeline()
indexing.add_component("sparse_embedder", sparse_embedder)
indexing.add_component("dense_embedder", dense_embedder)
indexing.add_component("writer", document_writer)
indexing.connect("sparse_embedder", "dense_embedder")
indexing.connect("dense_embedder", "writer")
logger.success("✨ Indexing pipeline created successfully")
return indexing
except Exception:
logger.exception("Failed to create indexing pipeline")
raise
async def create_query_pipeline(self) -> Pipeline:
"""Create query pipeline with concurrent component initialization"""
logger.info("Creating query pipeline...")
try:
# Initialize doc_store and text embedders concurrently
doc_store, (dense_embedder, sparse_embedder) = await asyncio.gather(
self._async_init(self._create_doc_store),
self._async_init(self._create_text_embedders),
)
# Initialize remaining components concurrently
retriever, reranker, generator, prompt_builder = await asyncio.gather(
self._async_init(partial(self._create_retriever, doc_store)),
self._async_init(self._create_reranker),
self._async_init(self._create_generator),
self._async_init(self._create_prompt_builder),
)
querying = Pipeline()
querying.add_component("sparse_embedder", sparse_embedder)
querying.add_component("dense_embedder", dense_embedder)
querying.add_component("retriever", retriever)
querying.add_component("reranker", reranker)
querying.add_component("prompt_builder", prompt_builder)
querying.add_component("llm", generator)
querying.connect(
"sparse_embedder.sparse_embedding", "retriever.query_sparse_embedding"
)
querying.connect("dense_embedder.embedding", "retriever.query_embedding")
querying.connect("retriever.documents", "reranker.documents")
querying.connect("reranker", "prompt_builder")
querying.connect("prompt_builder", "llm")
return querying
except Exception:
logger.exception("Failed to create query pipeline")
raise
FastAPI Application
During the start-up process of the FastAPI app, initial data will be loaded into the Qdrant vector database so that the RAG application itself can be utilized with immediate effect. However, before the data was loaded, the document store initializer must be spun up first to test the connection, ensuring that the Qdrant Docker container is fully functional before the API endpoints are made available to the users:
# app/main.py
from fastapi import FastAPI
from contextlib import asynccontextmanager
from app.api.endpoints import medication
from app.config.logging import get_logger
from app.config.settings import settings
from app.core.document_store.initializer import DocumentStoreInitializer
from app.core.initialization.data_loader import DataLoader
logger = get_logger(__name__)
@asynccontextmanager
async def lifespan(app: FastAPI):
"""Lifecycle manager for FastAPI application"""
logger.info("Initializing application components...")
initializer = DocumentStoreInitializer()
data_loader = DataLoader()
try:
# Test connection to document store
await initializer.test_connection()
if initializer._test_store is not None:
# Loads initial data
await data_loader.load_initial_data()
yield
except Exception:
logger.exception("Failed to initialize pipelines")
raise
finally:
logger.info("Shutting down application...")
app = FastAPI(
title=settings.PROJECT_NAME,
description=settings.PROJECT_DESCRIPTION,
lifespan=lifespan,
openapi_url=f"{settings.API_V1_STR}/openapi.json",
)
# Include routers
app.include_router(medication.router, prefix=settings.API_V1_STR, tags=["Medication"])
@app.get("/health", tags=["System"])
async def health_check():
"""Health check endpoint"""
return {"status": "healthy", "version": "1.0.0"}
The document store initializer uses a simple method to test the connection of the vector database by counting documents, though it can still fail when Qdrant is not finished with its start-up routine in the Docker container, so it is reliant on the @retry_with_logging decorator to automatically test the connection persistently in the event of failures:
# app/core/document_store/initializer.py
from typing import Optional
from haystack_integrations.document_stores.qdrant import QdrantDocumentStore
from app.core.document_store.factory import DocumentStoreFactory
from app.utils.retry import retry_with_logging
from app.config.logging import get_logger
logger = get_logger(__name__)
class DocumentStoreInitializer:
"""Handles initialization and testing of document store connection"""
def __init__(self):
self._test_store: Optional[QdrantDocumentStore] = None
@retry_with_logging
async def test_connection(self) -> None:
"""Test document store connection with retry logic"""
try:
factory = DocumentStoreFactory()
test_store = factory.create_document_store()
# Test the connection by performing a simple operation
_ = test_store.count_documents()
self._test_store = test_store
logger.info("Successfully tested Qdrant document store connection")
except Exception as e:
logger.error(f"Failed to connect to Qdrant: {str(e)}")
raise
async def cleanup(self) -> None:
"""Clean up test connection"""
if self._test_store:
try:
await self._test_store.client.close()
except Exception as e:
logger.error(f"Error during cleanup: {str(e)}")
The retry decorator logic is built on top of the Python package of tenacity, which is a general-purpose retrying library, one that is targeting the emergence of specific Qdrant HTTP exceptions to re-attempt retries.
# app/utils/retry.py
import logging
from functools import wraps
from qdrant_client.http.exceptions import UnexpectedResponse, ResponseHandlingException
from httpx import ConnectError, ReadTimeout, ConnectTimeout
from tenacity import (
retry,
stop_after_attempt,
wait_exponential,
retry_if_exception_type,
before_log,
after_log,
)
from app.config.logging import get_logger
logger = get_logger(__name__)
QDRANT_HTTP_EXCEPTIONS = (
ConnectionRefusedError,
ResponseHandlingException,
UnexpectedResponse,
ConnectError,
ConnectTimeout,
ReadTimeout,
)
def retry_with_logging(func):
@wraps(func)
async def wrapper(args, **kwargs):
try:
return await func(args, **kwargs)
except Exception as e:
logger.error(f"Failed to {func.__name__}: {str(e)}")
raise
retry_decorator = retry(
stop=stop_after_attempt(5),
wait=wait_exponential(multiplier=1, min=4, max=10),
retry=retry_if_exception_type(QDRANT_HTTP_EXCEPTIONS),
before=before_log(logger, logging.INFO),
after=after_log(logger, logging.INFO),
)
return retry_decorator(wrapper)
Asynchronous functions were employed to support concurrent requests for the API endpoints, there are two main endpoints that handles POST requests efficient:
- PATH: /extract
- Extracts entities from a given list of unstructured prescription text and output valid JSON format for disambiguated medication entities.
- PATH: /index
- Index medications into vector database, to add more few-shot examples for better in-context learning performance of the LLM.
# app/api/endpoints/medication.py
import traceback
from fastapi import APIRouter, Depends
from app.core.services.medication import MedicationService
from app.api.dependencies import get_medication_service
from app.config.logging import get_logger
from app.schemas.medication import (
MedicationRequest,
MedicationResponse,
MedicationIndexRequest,
MedicationIndexResponse,
)
logger = get_logger(__name__)
router = APIRouter()
@router.post("/extract", response_model=MedicationResponse)
async def extract_medications(
request: MedicationRequest,
medication_service: MedicationService = Depends(get_medication_service),
):
try:
result = await medication_service.extract_entities(request.texts)
return MedicationResponse(
results=result.results, processing_time=result.processing_time
)
except Exception as e:
logger.error(
f"An error was encountered while extracting entities: {e}.\n{traceback.format_exc()}"
)
raise
@router.post("/index", response_model=MedicationIndexResponse)
async def index_medications(
request: MedicationIndexRequest,
medication_service: MedicationService = Depends(get_medication_service),
):
"""
Index medication entities into the vector database for future retrieval.
"""
try:
result = await medication_service.index_medications(request.medications)
return MedicationIndexResponse(
message=result.message, processing_time=result.processing_time
)
except Exception as e:
logger.error(
f"An error was encountered while indexing medications: {e}.\n{traceback.format_exc()}"
)
raise
The API endpoint schemas are inclusive of JSON examples for better documentation and allowing users to examine the inputs and outputs of the respective endpoints.
# app/schemas/medication.py
from typing import List
from pydantic import BaseModel, Field
class MedicationEntity(BaseModel):
original_text: str = Field(..., description="Original medication text")
quantity: List[str] = Field(
default_factory=list, description="List of quantities found"
)
drug_name: List[str] = Field(
default_factory=list, description="List of drug names found"
)
dosage: List[str] = Field(default_factory=list, description="List of dosages found")
administration_type: List[str] = Field(
default_factory=list, description="List of administration types found"
)
brand: List[str] = Field(default_factory=list, description="List of brands found")
model_config = {
"json_schema_extra": {
"examples": [
{
"original_text": "Acetaminophen 325 MG Oral Tablet",
"quantity": [],
"drug_name": ["Acetaminophen"],
"dosage": ["325 MG"],
"administration_type": ["Oral Tablet"],
"brand": [],
}
]
}
}
class MedicationRequest(BaseModel):
texts: List[str] = Field(
...,
min_length=1,
max_length=100,
description="List of medication texts to process",
)
model_config = {
"json_schema_extra": {
"examples": [{"texts": ["Acetaminophen 325 MG Oral Tablet"]}]
}
}
class MedicationResponse(BaseModel):
results: List[MedicationEntity] = Field(
..., description="List of extracted medication entities"
)
processing_time: float = Field(..., description="Total processing time in seconds")
model_config = {
"json_schema_extra": {
"examples": [
{
"results": [
{
"original_text": "Acetaminophen 325 MG Oral Tablet",
"quantity": [],
"drug_name": ["Acetaminophen"],
"dosage": ["325 MG"],
"administration_type": ["Oral Tablet"],
"brand": [],
}
],
"processing_time": 0.15,
}
]
}
}
class MedicationIndexRequest(BaseModel):
medications: List[MedicationEntity] = Field(
..., min_length=1, description="List of medications to index"
)
class MedicationIndexResponse(BaseModel):
message: str = Field(
..., description="Message indicating the success or failure of the operation"
)
processing_time: float = Field(..., description="Total processing time in seconds")
model_config = {
"json_schema_extra": {
"examples": [
{
"message": "Successfully indexed 1 entity",
"processing_time": 0.05,
}
]
}
}
Services
A microservice-like architecture was adopted to construct the services that the RAG application is dependent on, to allow flexibility and modularity in the codebase so that future addition of features and updates can be integrated with ease. Currently, there are two existing services: MedicationService and PipelineService. MedicationService handles the logic behind the POST request of both the “/extract” and “/index” endpoints, while PipelineService handles the lifecycle logic of instantiating either the indexing or querying pipelines when it is invoked by MedicationService.
MedicationService
# app/core/services/medication.py
import json
import time
import uuid
from typing import List, Dict, Any
from app.core.services.pipeline import PipelineService
from app.utils.common import create_index_documents
from app.schemas.medication import (
MedicationEntity,
MedicationResponse,
MedicationIndexResponse,
)
from app.config.logging import get_logger
logger = get_logger(__name__)
class MedicationService:
"""Service for processing medication-related operations"""
def __init__(self, pipeline_service: PipelineService):
self._pipeline_service = pipeline_service
async def index_medications(
self, medications: List[MedicationEntity]
) -> MedicationIndexResponse:
"""
Index medication entities into the vector database.
Args:
medications: List of medication entities to index
Returns:
IndexingResult containing indexing operation metadata
Raises:
IndexingError: If indexing operation fails
"""
request_id = str(uuid.uuid4())
logger.info(
f"Starting indexing operation for request {request_id} "
f"with {len(medications)} medications"
)
start_time = time.perf_counter()
try:
# Convert medication entities to indexable documents
documents = create_index_documents(medications)
# Execute indexing pipeline
await self._pipeline_service.execute_index_pipeline(documents)
processing_time = time.perf_counter() - start_time
logger.info(
f"Request {request_id}: Successfully indexed {len(medications)} "
f"medications in {processing_time:.2f} seconds"
)
return MedicationIndexResponse(
message=f"Successfully indexed {len(medications)} medications",
processing_time=processing_time,
)
except Exception as e:
logger.exception(f"Request {request_id}: Failed to index medications. {e}")
raise
async def extract_entities(self, texts: List[str]) -> MedicationResponse:
"""
Extract medication entities from a list of texts.
Args:
texts: List of medication strings to process
Returns:
ProcessingResult containing extracted entities and metadata
Raises:
EntityExtractionError: If extraction fails
"""
request_id = str(uuid.uuid4())
logger.info(f"Starting entity extraction for request {request_id}")
start_time = time.perf_counter()
results: List[MedicationEntity] = []
try:
for idx, text in enumerate(texts, 1):
logger.debug(
f"Request {request_id}: Processing text {idx}/{len(texts)}: {text}"
)
# Execute pipeline and extract entities
entities = await self._process_single_text(text, request_id, idx)
results.append(entities)
processing_time = time.perf_counter() - start_time
logger.info(
f"Request {request_id}: Completed processing {len(texts)} texts "
f"in {processing_time:.2f} seconds"
)
return MedicationResponse(results=results, processing_time=processing_time)
except Exception as e:
logger.exception(
f"Request {request_id}: Error during entity extraction. {e}"
)
raise
async def _process_single_text(
self, text: str, request_id: str, idx: int
) -> MedicationEntity:
"""Process a single medication text and extract entities"""
try:
# Execute query pipeline
response = await self._pipeline_service.execute_query_pipeline(text)
# Parse LLM response
extracted_data = self._parse_llm_response(response, text)
logger.debug(
f"Request {request_id}: Successfully extracted entities from text {idx}"
)
return MedicationEntity(**extracted_data)
except Exception as e:
logger.error(
f"Request {request_id}: Failed to process text {idx}: {str(e)}"
)
# Return empty entity on failure
return MedicationEntity(original_text=text)
def _parse_llm_response(
self, llm_response: str, original_text: str
) -> Dict[str, Any]:
"""Parse LLM response into structured data"""
try:
extracted = json.loads(llm_response["llm"]["replies"][0])
extracted["original_text"] = original_text
return extracted
except json.JSONDecodeError as e:
logger.error(f"Failed to parse LLM response: {str(e)}")
return {
"original_text": original_text,
"quantity": [],
"drug_name": [],
"dosage": [],
"administration_type": [],
"brand": [],
}
PipelineService
# app/core/services/pipeline.py
import traceback
from time import perf_counter
from typing import List, Dict, Union, Any
from contextlib import asynccontextmanager
from pydantic import BaseModel
from haystack import Pipeline
from haystack.dataclasses import Document
from app.core.pipeline.factory import PipelineFactory
from app.config.logging import get_logger
logger = get_logger(__name__)
class PipelineMetrics(BaseModel):
"""Metrics for pipeline execution"""
pipeline_creation_time: float
execution_time: float
total_time: float
class PipelineService:
"""Service for managing and executing pipelines"""
def __init__(self, pipeline_factory: PipelineFactory):
self._pipeline_factory = pipeline_factory
@asynccontextmanager
async def _pipeline_lifecycle(self, pipeline_type: str):
"""Manage pipeline lifecycle and measure performance"""
start_time = perf_counter()
pipeline = None
try:
if pipeline_type == "query":
pipeline = await self._pipeline_factory.create_query_pipeline()
elif pipeline_type == "index":
pipeline = await self._pipeline_factory.create_indexing_pipeline()
else:
raise ValueError(f"Unknown pipeline type: {pipeline_type}")
creation_time = perf_counter() - start_time
logger.debug(
f"{pipeline_type.capitalize()} pipeline created in {creation_time:.2f}s"
)
try:
yield pipeline, creation_time, start_time
finally:
# Cleanup could go here if needed
pass
except Exception as e:
logger.error(f"Pipeline lifecycle error: {str(e)}")
raise
async def execute_query_pipeline(self, text: str) -> Dict[str, Any]:
"""Execute query pipeline with fresh components"""
if not isinstance(text, str) or not text.strip():
raise ValueError("Query text must be a non-empty string")
async with self._pipeline_lifecycle("query") as (
pipeline,
creation_time,
start_time,
):
try:
pipeline_input = self._create_query_input(text)
result = await self._run_pipeline(pipeline, pipeline_input)
# Calculate and log metrics
metrics = self._calculate_metrics(creation_time, start_time)
logger.info(
"Query pipeline metrics: "
f"creation={metrics.pipeline_creation_time:.2f}s, "
f"execution={metrics.execution_time:.2f}s, "
f"total={metrics.total_time:.2f}s"
)
return result
except Exception as e:
logger.error(
f"Query pipeline execution failed: {str(e)}",
extra={"query_text": text[:100]}, # Log truncated query for context
)
raise
async def execute_index_pipeline(
self, documents: List[Union[Dict, Document]]
) -> None:
"""Execute indexing pipeline with fresh components"""
if not documents:
raise ValueError("Documents list cannot be empty")
# Convert dict documents to Document objects if needed
processed_docs = self._prepare_documents(documents)
async with self._pipeline_lifecycle("index") as (
pipeline,
creation_time,
start_time,
):
try:
pipeline_input = self._create_index_input(processed_docs)
await self._run_pipeline(pipeline, pipeline_input)
# Calculate and log metrics
metrics = self._calculate_metrics(creation_time, start_time)
logger.info(
f"Indexed {len(documents)} documents - "
f"creation={metrics.pipeline_creation_time:.2f}s, "
f"execution={metrics.execution_time:.2f}s, "
f"total={metrics.total_time:.2f}s"
)
except Exception as e:
logger.error(
f"Index pipeline execution failed: {str(e)}",
extra={"document_count": len(documents)},
)
raise
@staticmethod
def _create_query_input(text: str) -> Dict[str, Dict[str, str]]:
"""Create formatted input for query pipeline"""
return {
"sparse_embedder": {"text": text},
"dense_embedder": {"text": text},
"reranker": {"query": text},
"prompt_builder": {"query": text},
}
@staticmethod
def _create_index_input(
documents: List[Document],
) -> Dict[str, Dict[str, List[Document]]]:
"""Create formatted input for index pipeline"""
return {"sparse_embedder": {"documents": documents}}
async def _run_pipeline(
self, pipeline: Pipeline, pipeline_input: Dict[str, Any]
) -> Dict[str, Any]:
"""Execute pipeline with error handling"""
try:
return pipeline.run(pipeline_input)
except Exception as e:
logger.error(
f"Pipeline run failed: {str(e)}.\n{traceback.format_exc()}",
extra={
"pipeline_input": str(pipeline_input)[:200]
}, # Log truncated input
)
raise
@staticmethod
def _prepare_documents(documents: List[Union[Dict, Document]]) -> List[Document]:
"""Convert dictionary documents to Document objects if needed"""
processed_docs = []
for doc in documents:
if isinstance(doc, dict):
processed_docs.append(Document(**doc))
elif isinstance(doc, Document):
processed_docs.append(doc)
else:
raise ValueError(
f"Invalid document type: {type(doc)}. "
"Must be either dict or Document"
)
return processed_docs
@staticmethod
def _calculate_metrics(creation_time: float, start_time: float) -> PipelineMetrics:
"""Calculate pipeline execution metrics"""
current_time = perf_counter()
return PipelineMetrics(
pipeline_creation_time=creation_time,
execution_time=current_time - start_time - creation_time,
total_time=current_time - start_time,
)
Evaluation
The evaluation of the RAG app is based on the evaluation dataset where the medication texts “original_text” are processed, and the validity of the JSON outputs produced by the LLM, are compared against the one found in the dataset which serves as ground truth. Therefore, the evaluation metrics such as accuracy, precision, recall, and f1-score are measured based on binary labels (i.e. “1” = Correct, “0” = Wrong), whether the JSON output matches to that of the ground truth equivalent.
# app/core/evaluation/evaluator.py
import json
from time import perf_counter
from pydantic import BaseModel
from typing import Dict, Any, List, Tuple
from app.core.pipeline.factory import PipelineFactory
from app.schemas.medication import MedicationEntity
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from app.config.logging import get_logger
logger = get_logger(__name__)
class EvaluationOutput(BaseModel):
accuracy: float
precision: float
recall: float
f1_score: float
class Evaluator:
def __init__(self):
self._factory = PipelineFactory()
def _format_llm_response(self, response: Dict[str, Any]) -> Tuple[str, List[str]]:
"""Format the LLM response into usable format for evaluation."""
try:
answer = json.loads(response["llm"]["replies"][0])
_contexts = [doc.meta for doc in response["reranker"]["documents"]]
contexts = [
f"Query: {ctx["original_text"]}\nAnswer: {ctx}" for ctx in _contexts
]
return answer, contexts
except Exception as e:
logger.error(f"Error formatting LLM response: {e}")
raise
def _compute_metrics(self, dataset: Dict[str, float]) -> EvaluationOutput:
"""Compute standard ML evaluation metrics for the test dataset."""
try:
generated_answer = dataset["answer"]
ground_truth = dataset["ground_truth"]
# Compare JSON objects and output True/False
boolean_comparisons = [
gen == gt for gen, gt in zip(generated_answer, ground_truth)
]
# Convert boolean comparisons to binary labels (True -> 1, False -> 0)
y_pred = [int(b) for b in boolean_comparisons]
y_true = [1] * len(
generated_answer
) # Since ground truth is considered "correct", it is all 1s
# Compute metrics
accuracy = accuracy_score(y_true, y_pred)
precision = precision_score(y_true, y_pred, zero_division=0)
recall = recall_score(y_true, y_pred, zero_division=0)
f1 = f1_score(y_true, y_pred, zero_division=0)
return EvaluationOutput(
accuracy=accuracy, precision=precision, recall=recall, f1_score=f1
)
except Exception as e:
logger.error(f"Error during evaluation: {e}")
return EvaluationOutput(
accuracy=None, precision=None, recall=None, f1_score=None
)
async def run(self, test_data: List[MedicationEntity]) -> EvaluationOutput:
"""Run the evaluation on the provided test dataset."""
eval_dataset = {"question": [], "answer": [], "context": [], "ground_truth": []}
try:
start = perf_counter()
for _input in test_data:
query = _input.original_text
query_pipeline = await self._factory.create_query_pipeline()
llm_response = query_pipeline.run(
data={
"sparse_embedder": {"text": query},
"dense_embedder": {"text": query},
"reranker": {"query": query},
"prompt_builder": {"query": query},
},
include_outputs_from={"reranker"},
)
answers, contexts = self._format_llm_response(llm_response)
eval_dataset["question"].append(query)
eval_dataset["answer"].append(answers)
eval_dataset["context"].extend(contexts)
eval_dataset["ground_truth"].append(_input.model_dump())
elapsed = perf_counter() - start
logger.info("Evaluation completed successfully.")
logger.info(
f"Evaluation took {elapsed:.2f} seconds for processing {len(test_data)} medication text."
)
return self._compute_metrics(eval_dataset)
except Exception as e:
logger.error(f"Error during evaluation: {e}")
raise
Docker Containerization
The entire application is orchestrated with Docker, as it can be easily configured and deployed on any compute instances. The configurations are highly customizable, allowing users to inject preferred settings in the form of environment variables according to their specific use cases. The Ollama service requires a GPU to run the RAG app seamlessly, but it can be modified to utilize only the CPU to perform inferences. More details can be found in the GitHub repo.
# docker-compose.yml
services:
app:
build:
context: .
dockerfile: docker/app/Dockerfile
container_name: app
image: ner-rag-app:v1.0.0
environment:
- HAYSTACK_TELEMETRY_ENABLED=False
env_file:
- .env
depends_on:
- qdrant
- ollama
ports:
- ${FASTAPI_PORT}:8000
networks:
- rag-app-network
qdrant:
image: qdrant/qdrant:latest
restart: always
container_name: qdrant
env_file:
- .env
ports:
- ${QDRANT_PORT_EXTERNAL}:6333
- 6334:6334
volumes:
- qdrant_data:/qdrant/storage
networks:
- rag-app-network
ollama:
image: ollama/ollama:latest
restart: always
container_name: ollama
env_file:
- .env
environment:
- OLLAMA_HOST=0.0.0.0
volumes:
- ollama_data:/root/.ollama
- ./docker/ollama:/scripts
ports:
- ${OLLAMA_API_PORT_EXTERNAL}:11434
entrypoint: ["/bin/bash", "/scripts/entrypoint.sh"]
networks:
- rag-app-network
tty: true
# pull_policy: always
deploy:
resources:
reservations:
devices:
- driver: nvidia
count: 1
capabilities: [gpu]
volumes:
qdrant_data:
ollama_data:
networks:
rag-app-network:
driver: bridge
Performance
Tested on an evaluation dataset consisting of 335 medication texts.
Entity Extraction
- Accuracy: 92.2 %
- Precision: 100.0 %
- Recall: 92.2 %
- F1 Score: 95.9 %
Latency
- Average processing time per medication text: 1.33 seconds
As you might have guess, this RAG app is not suitable for real-time applications due to its latency. However, it can be used in batch processing or as a background task in larger systems.
Conclusion
This project demonstrates how a lightweight RAG system can solve a practical healthcare problem especially in the NLP domain. By combining efficient LLMs, hybrid search, and RAG systems, it’s possible to unlock the value of unstructured medication data at scale.
Whether you’re a developer, data scientist, or healthcare professional, I hope this inspires you to tackle similar challenges in your domain. Let’s continue building tools that make healthcare smarter and more efficient!
Reference
- Retrieval Augmented Generation (RAG) Explained (Source)
- Rerankers and Two-Stage Retrieval (Source)
- Less is More: How Good RAG Design Lets You Use Smaller Language Models (Source)
Icon Attribution
RAG Architecture Diagram
- Document —> Author: manshagraphics [Flaticon]
- Vectors —> Author: Triangle Squad [Flaticon]
- User —> Author: Smashicons [Flaticon]
- Query —> Author: Freepik [Flaticon]
- Hybrid Search —> Author: Vitaly Gorbachev [Flaticon]
- Reranker —> Author: Freepik [Flaticon]
- Response —> Author: Ilham Fitrotul Hayat [Flaticon]
